The IMPC is increasing our understanding of the genetic basis for metabolic diseases
- Metabolic diseases, such as obesity and diabetes, affect people worldwide
- The function of many genes in the genome is still unknown
- Knockout mice allow us to understand metabolic procedures and relate them to human disease
Approach
To identify the function of genes, the IMPC uses a series of standardised protocols described in IMPReSS (International Mouse Phenotyping Resource of Standardised Screens). Tests addressing the metabolic function are conducted on young adult mice at 11-16 weeks of age.
Procedures that can lead to relevant phenotype associations
Young Adult:- Calorimetry: ESLIM v1,
- Challenge Whole Body Plethysmography: TCP v3, ICS v3,
- Clinical Blood Chemistry: HAS v2,
- Clinical Chemistry: HRWLLA v2, ICSLA v3, HAROLD v1, HRWLLA v3, IMPC v3, IMPC v3, IMPC v1, IMPC v1, IMPC v2, IMPC v2, UCDLA v3, IMPC v2, HMGULA v2, IMPC v3, JAXLA v3, IMPC v3, IMPC v3, IMPC v2, KMPCLA v3, IMPC v1, IMPC v3, BCMLA v3, IMPC v3, M-G-P v1, TCPLA v3, RBRCLA v3, IMPC v2, IMPC v3, IMPC v1, ESLIM v1, IMPC v1, IMPC v2, NINGLA v3, IMPC v2, IMPC v2, IMPC v1, IMPC v1, HMGULA v2, IMPC v2, IMPC v3, IMPC v3, IMPC v1,
- Clinical chemistry (GMC): GMC v1,
- Echo: HMGULA v1, HRWLLA v1, HMGULA v1, IMPC v1, IMPC v1, IMPC v1, IMPC v1, IMPC v1, NINGLA v1, BCMLA v1, IMPC v1, ICSLA v1,
- Fast Bleed: HAS v1,
- Fasted Clinical Chemistry: ESLIM v1,
- Food efficiency (GMC): GMC v1,
- IPGTT: M-G-P v1,
- Indirect Calorimetry: IMPC v1, IMPC v3, IMPC v2, HAS v2, IMPC v3, IMPC v2, IMPC v1, IMPC v3, IMPC v2, M-G-P v1, IMPC v1, IMPC v2, IMPC v1, HMGULA v3, IMPC v2, IMPC v3, IMPC v3, IMPC v1, IMPC v3, IMPC v2, IMPC v2, NINGLA v3, HMGULA v3, KMPCLA v3, ICSLA v3, IMPC v2, IMPC v3, IMPC v1, BCMLA v3, IMPC v3, RBRCLA v3, IMPC v1, IMPC v3,
- Insulin Blood Level: IMPC v3, IMPC v3, IMPC v2, IMPC v1, IMPC v1, IMPC v3, IMPC v1, IMPC v2, IMPC v3, IMPC v2, IMPC v2, IMPC v2, KMPCLA v3, IMPC v3, IMPC v1, IMPC v3, IMPC v2, IMPC v1, IMPC v1, IMPC v2, IMPC v1, HMGULA v3, IMPC v1, IMPC v3, HMGULA v3, IMPC v3, HAS v2, IMPC v2,
- Insulin t=0: HAROLD v1,
- Intraperitoneal glucose tolerance test (IPGTT): ICSLA v1, IMPC v1, IMPC v1, HMGULA v1, UCDLA v1, RBRCLA v1, MGP v1, IMPC v1, KMPCLA v1, BCMLA v1, IMPC v1, NINGLA v1, HRWLLA v1, IMPC v1, HMGULA v1, IMPC v1, HAS v2, TCPLA v1, IMPC v1, IMPC v1, JAXLA v1, IMPC v1, IMPC v1,
- MicroCT E14.5-E15.5: IMPC v2, IMPC v2, IMPC v2, IMPC v2, IMPC v2, IMPC v2, IMPC v2,
- MicroCT E18.5: IMPC v2, IMPC v2, IMPC v2, IMPC v2, IMPC v2, IMPC v2,
- Plasma Chemistry: JAX v1,
- Simplified IPGTT: ESLIM v1, HAROLD v1,
- Terminal Bleed: HAS v1,
- Urinalysis: JAX v1,
- Urinalysis: HAS v1,
IMPC Metabolism Publication
Metabolic diseases investigated in 2,016 knockout mouse lines
Nature Communications publication
- 974 genes were identified with strong metabolic phenotypes (see gene table, below).
- 429 genes had not been previously associated with metabolism, 51 completely lacked functional annotation, and 25 had single nucleotide polymorphisms associated to human metabolic disease phenotypes.
- 515 genes linked to at least one disease in OMIM.
- Networks of co-regulated genes were identified, and genes of predicted metabolic function found.
- Pathway mapping revealed sexual dimorphism in genes and pathways.
- This investigation is based on about 10% of mammalian protein-coding genes. The IMPC will continue screening for genes associated to metabolic diseases in its second 5 year phase.
Gene table
Mutant/wildtype ratios below the 5th percentile and above the 95th percentile of the ratio distributions yielded 28 gene lists that serve as a data mining resource for further investigation into potential links to human metabolic disorders.
By hovering over the table you can select cells and click to explore the underlying data.
| Outlier | ||||
| <5% | >95% | |||
| Fasting Glucose (T0) | 94 | 96 | 96 | 96 |
| Glucose Response (AUC) | 96 | 96 | 96 | 96 |
| Triglycerides (TG) | 72 | 72 | 72 | 72 |
| Body Mass (BM) | 86 | 86 | 86 | 86 |
| Metabolic Rate (RM) | 18 | 48 | 18 | 48 |
| Oxygen Consumption Rate (V02) | 18 | 48 | 18 | 48 |
| Respiratory Exchange Ratio (RER) | 17 | 47 | 17 | 47 |
Strong metabolic phenotype genes form regulatory networks
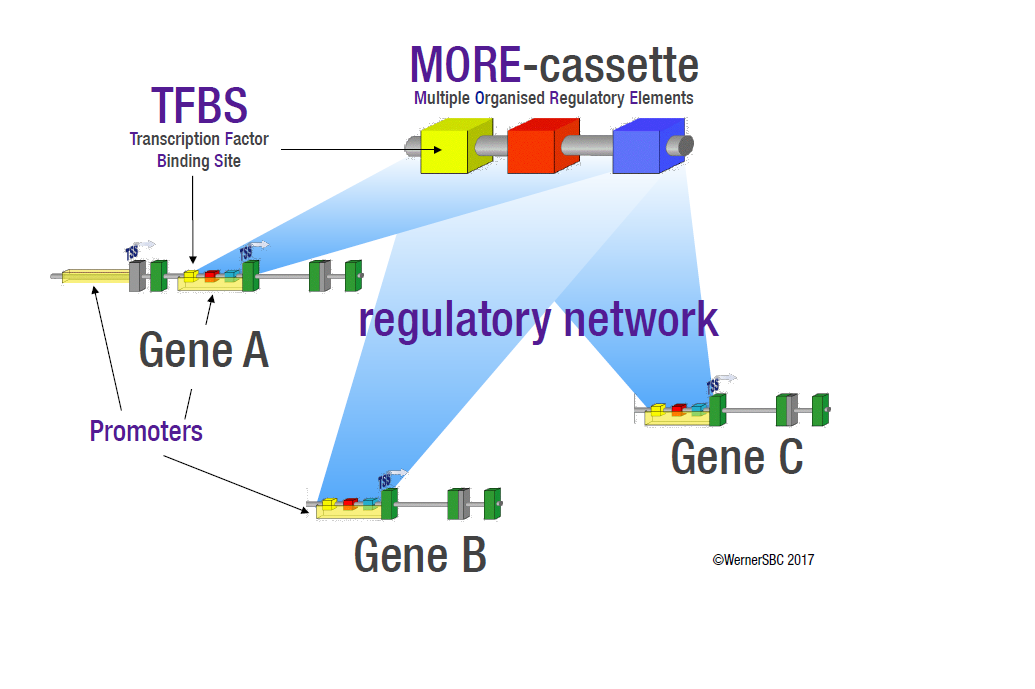
- Transcriptional co-regulation often involves a common set of transcription factor binding sites (TFBSs) shared between co-regulated promoters and in a particular organization (known as Multiple Organized Regulatory Element (MORE)–cassettes).
- Identification of shared MORE-cassettes in promoters of candidate genes allowed to discover extensive metabolic phenotype-associated networks of potentially co-regulated genes.
- MORE-cassettes are invariant genomic sequence features (similar to reading frames).
- The presence of MORE-cassettes enabled to a priori predict phenotypes and identify genes potentially linked to metabolic functions.
Methods
Genes with phenotypes associated to the following seven metabolic parameters, with diagnostic relevance in human clinical research, were further analysed:
- Fasting basal blood glucose level before glucose tolerance test (T0)
- Area under the curve of blood glucose level after intraperitoneal glucose administration relative to basal blood glucose level (AUC)
- Plasma triglyceride levels (TG)
- Body mass (BM)
- Metabolic rate (MR)
- Oxygen consumption rate (VO2)
- Respiratory exchange ratio (RER) – a measure of whole-body metabolic fuel utilization
Mutant/wildtype ratios (mean value of metabolic parameters of mutants divided by the mean value obtained for wildtypes) were calculated:
- Control wildtype mice from each phenotypic center included, matched for sex, age, phenotypic pipeline and metadata (e.g. instrument).
- Males and females analyzed separately to account for sexual dimorphism.
New mouse models
- IMPC generated and identified new genetic disease models.
- New models available to the research community to perform in-depth investigations of novel genetic elements associated to metabolic disease mechanisms.
- These models fill the gap between genome-wide association studies and functional validation using a mammalian model organism.
Phenotypes distribution
Homeostasis/metabolism IKMC/IMPC related publications
Showing 1 to 0 of 0 entries
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |