Researchers at the Jackson Laboratory, an IMPC consortium member, have created a new automated scoring system for nocifensive behaviour. Pain studies usually have to be done manually by researchers, making them slow and labour intensive. An automated scoring system can increase the speed and reliability of measurements, thus producing beneficial impacts from pain research more quickly.
Nocifensive Behaviour
Pain plays an important protective role for humans and other organisms. It informs us of injuries and illnesses, whether it be a skin infection, viral cough or a broken bone. Genetic mutations can cause reduced or absent sensitivity to pain, leading to frequent injuries and high mortality rates.
Animal models are crucial for studying relationships between genes and pain responses. Pain in mice is expressed as ‘nocifensive behaviours’, behavioural responses that protect against injury in response to pain, such as licking or limping. Wotton et al. created an automated scoring system using machine learning methods that detects and records licking/biting behaviour in the formalin assay. As a test, the system was used to compare C57BL/6J mice to C57BL/6NJ mice.
The Formalin Test
Researchers use known and well-practised methods for measuring pain in animal models. Formalin is a chemical irritant that causes localised inflammation. It is usually injected into a hind paw, causing nocifensive behaviour such as licking, biting, lifting, flicking or clutching of the paw.
Typically in formalin tests, the mice are videoed over the time course of the response with individual researchers carefully watching and recording nocifensive behaviours. This makes the formalin assay labour-intensive and open to observer variability or human error. An hour of video of a single mouse usually takes up to two hours to fully score.
An automated scoring system of video recordings could solve many of these issues. It would be easy to implement, inexpensive, accessible, and causes no additional stress to the mice. Machine learning is a rapidly advancing area, with recent developments allowing systems to detect tiny differences in mouse facial expressions and other behaviour. Licking and biting are clear and easy to label behaviours, well-suited for machine learning classification.
Integrating a machine learning system with the formalin assay would not only save on time and labour. It would also provide scalability, reliability and reproducibility. It could be a crucial technological jump for chronic pain research and related areas.
Training the Model
The mice were anesthetised before formalin was injected into the right hind paw to reduce stress and maximise consistency. They were then placed in the testing arena where they were videoed for 90 minutes.
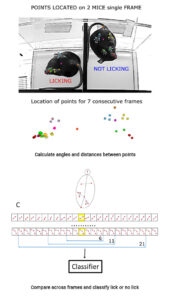
The system model was divided into three separate modules, the first being “key point detection” – learning how to track the different body parts of mice. To do this, the researchers created a ‘training set’ by videoing mice and manually labelling each frame. Each mouse was labelled with 12 points: mouse, nose, right front paw, left front paw, 3 points on each hind paw (outer, inner, base), mid-abdomen and tail base. The testing arena was also labelled with 5 points, making it 53 points per frame. The system’s point tracker needed to learn to recognise all 53 points. Empty arenas and different lighting conditions were used for variability. Wotton et al. used a software tracker call DeepLabCut, enabling the system to estimate the poses and movements of the mice.
The second module was “per frame feature extraction” – using the pixel coordinates and probability of each key point to classify behaviour. The system was able to ‘try and find’ each key point and estimated the likelihood that it was correct in its detection. Several different angles and distances were used for each body part. Using angles and distances to estimate the position of body parts is a low-cost way to generate pose features per frame.
Lastly, the third module was “behaviour classification” – where the model took the frame-based features it had learned in the first two modules and classified each frame with a behaviour, such as licking or biting.
Initial Validation
After testing the classifier on the training videos, Wotton et al. found the automated model had a 98% accuracy compared to humans. Forty-three of the videos had no licking behaviour, the human and model had an average agreement of 98.8%, suggesting a low false-positive rate. Two videos out of the total 111 had a lower average agreement, at around 84%. Closer inspection of these clips showed the mice had ambiguous behaviour, making it difficult for the human researcher to determine if there was licking behaviour.
Wotton et al. also tested the model’s ability to differentiate between levels of behaviour, such as no licking, some licking or a lot of licking. For each of the 111 video clips, the length of licking behaviour was determined by a human observer and the videos were ranked. The clips were then grouped based on similar levels of behaviour, with each of the groups having a 20-second mean difference to each other.
The human observer and the model could easily differentiate between these groups due to the 20-second difference in behaviour length. Wotton et al., therefore, shifted the time difference between groups to determine the smallest difference that could be reliably detected. For the human observers, this was a difference of 11 seconds whilst the model needed a difference of 16 seconds.
The researchers concluded that the model could reliably distinguish biologically realistic differences of 13% for licking/biting behaviour. This would be enough to detect known differences between mouse strains, such as the two inbred strains (C57BL/6J and C57BL/6NJ) in this study.
Inter-observer validity was also tested with a second observer. A high level of agreement between the two observers and the model was found.
Strain Comparison
Wotton et al. decided to test the accuracy of the model by getting it to detect an already known difference between the two mouse model strains. Bryant et al. previously manually used the formalin test to compare C57BL/6NCrl and C57BL/6J mice. They found that male C57BL/6NCrl mice had a reduced licking response in phase II of the assay (20-45 minutes after formalin injection) There was no significant difference for females.
The mouse strains Wotton et al. used are very similar to the two used in the Bryant et al. study. Wotton et al. found that male C57BL/6NJ mice showed reduced licking compared to C57BL/6J male mice. It was the reverse for the female mice, with C57BL/6NJ females licking more. The researchers also found that these sex/strain differences were more easily visible when analysing data for the full 90 minutes of the formalin assay instead of just the 20-45 minute time span.
Bootstrapped statistical analysis of two different bin sizes (a 20-45 minute bin and a 10-60 minute bin) with an increasing sample size was conducted. For the 20-45 bin, it was found that “as sample size increases, the probability of finding a significant difference between the strains also increases, for both males and females.” This was not the case for the 10-60 bin, with the larger time span for observation minimising differences in females and suggesting that, contrary to the initial conclusion, there is no strain difference for female mice. This was not the case for the male mice: “the male probability of detecting a difference increases with sample size, the males appear to differ in the amount of licking, in both the amplitude and duration of peak behaviour.”
“Both the automated system and Bryant et al. showed that C57BL/6N males lick less regardless of the bin choice, but bin size for females heavily influenced the outcome.”
The choice of bin size for analysis can therefore have a large impact on the conclusions drawn from data.
The researchers concluded that licking behaviour was highly variable or, put more simply, “some mice lick more than others.” This highlights how the formalin assay is less likely to find a significant difference if using small numbers of mice.
Impacts
Wotton et al. concluded that the automated model system’s ability to classify behaviour was very comparable to humans.
“[It] had approximately 98% agreement with a human observer on a second by second basis, and it was also highly correlated with bin scoring over both long and short videos with two human observers.”
The automated system took an average of 2 hours to score four mice instead of 9 hours for a human researcher. The system can also analyse multiple videos simultaneously for 24 hours a day, seven days a week! Different system components, such as window size or classifier, could be easily substituted to suit each researcher and/or study. Larger sample sizes and long videos would normally incur larger annotation costs but using an automated system could eliminate this.
Wotton et al. showed that there are differences between mouse strains and sexes for formalin test data. They also emphasised that many other factors could affect formalin test data, including “environment, dose, injection quantity, anaesthesia, site of injection, time of day, experimenter effects, and observer bias.”
Systems that enable high-throughput pain studies are important for our scientific understanding. With mice still making up the vast majority of animal model use, it is vital that researchers understand strain or sex differences when choosing a model. In some cases, an automatic system can eliminate factors that cause differences, such as observer bias. In other cases, it allows for larger studies to be more easily conducted and for more data to be produced. This makes it easier for researchers to study the effects of factors on nocifensive behaviour (and other data.)
The impacts of this system also reach beyond screening for differences between mouse strains. The automated scoring system improves the reliability and speed of data production for all studies involving nocifensive behaviour. This includes preclinical studies for new drugs and therapies or studies targeting specific human diseases that cause chronic pain. More progress in these areas would have a valuable impact on human health.
For more information on the IMPC pain data, see our Pain Data Summary page.
Source:
Wotton, J. M. et al. (2020) ‘Machine learning-based automated phenotyping of inflammatory nocifensive behavior in mice’, Molecular Pain. doi: 10.1177/1744806920958596.