Data integration
The IMPC is seeking to identify genes that are critical for development and health and, ultimately, associated to human disease.
IMPC researchers have proposed a gene classification system involving cross comparing viability and phenotyping data from knockout IMPC mice with human cell essentiality scores from the Cancer Dependency map. The classification, called Full Spectrum of Intolerance to Loss-of-function (FUSIL), comprises five mutually exclusive bins, which range from more to less essential. Genes are ascertain to a FUSIL bin, which can be used to identify genes associated with disease. In this way, genes can be categorised as to how essential they are for supporting life and the likelihood they are associated with de novo genetic disorders.
This process requires integrating data collected from different sources (mouse gene identifiers, human gene identifiers, orthologue identification, IMPC viability, Achilles gene effect) to derive the FUSIL categories.
The databases which are the source of the data keep growing, as more data is generated and integrated. Thus, the IMPC has created a webapp tool that can achieve this automatically, the Essential Genes Data portal. In addition to the above mentioned gene attributes, we integrate gnomAD constraint scores, ClinGen haploinsufficiency data and IDG categories. Please continue reading for more information.
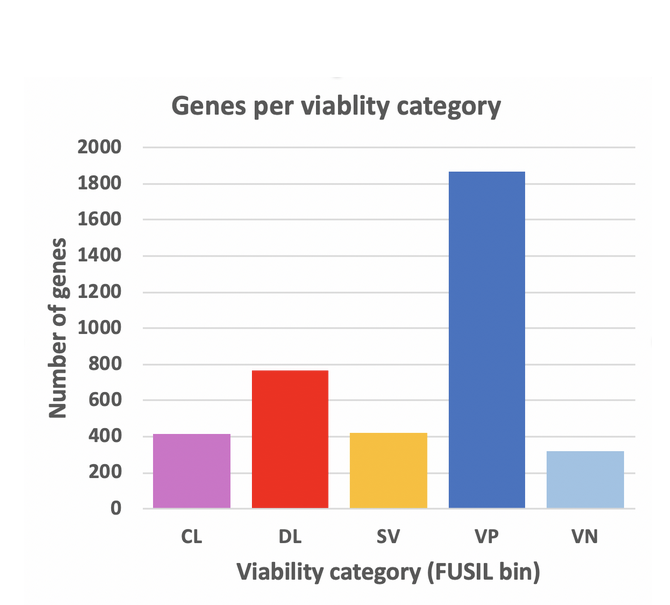
A Full Spectrum of Intolerance to Loss of Function (or FUSIL) score was developed to classify genes in bins of more to less essentiality:
- CL- Cellular Lethal
- DL- Developmental Lethal
- SV- Subviable
- VP- Viable with significant phenotype(s)
- VN- Viable with no signficant phenotypes
Genes associated with developmental disorders were overrepresented among genes in the DL bin.
