Emory scientists have identified a function for a mysterious DNA modification in fruit flies’ brain development, which may provide hints to its role in humans. The results are published in Molecular Cell.
Epigenetics may mean “above the genes,” but a lot of the focus in the field is on DNA methylation, a chemical modification of DNA itself. Methylation doesn’t change the actual DNA letters (A, C, G and T), but it does change how DNA is handled by the cell. Generally, it shuts genes off and is essential for cell differentiation.
The most commonly studied form of DNA methylation appears on the DNA letter C (cytosine). Drosophila, despite being a useful genetic model of development, have very little of this form of DNA methylation. What they do have is methylation on A (technically, N6-methyladenine), although little was known about what this modification did for flies.
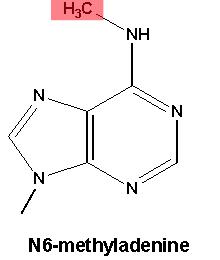
This is N6-methyladenine. Credit: Emory University.
Emory geneticists Bing Yao, PhD, Peng Jin, PhD and colleagues now have shown that an enzyme that removes methylation from A is critical for neuronal development in Drosophila.
This finding is significant because the enzyme is in the same family (TET for ten-eleven translocation) of demethylases that trigger removal of DNA methylation from C in mammals. The function of TET enzymes, revealing that cells actively removed DNA methylation rather than just letting it slough off, was discovered only in 2009.
From the point of view of the proteins that need to recognize it, methylation is really just a bump on a long DNA molecule, and that bump appears in a different context on adenine versus cytosine. Thus, it is striking that the enzymes that remove methylation are related in mammals and Drosophila.
Yao is assistant professor of human genetics, and Jin is vice chair of research in the Department of Human Genetics. They published a paper last year in Nature Communications showing that N6-methyladenine is present in the mouse genome and its levels in brain cells change in response to stress. Another lab recently reported that in the human genome, DNA methylation on adenine is extensive, if rare (1 in 2000 adenines), and reduced in cancer cells.
The fly enzyme that removes N6-methyladenine is called DMAD (DNA N6-methyladenine demethylase). Flies with mutations eliminating the DMAD gene cannot survive early development. The Emory researchers created flies that shut off DMAD in the brain, which perturbed development of their mushroom bodies, involved in learning and memory.
In fly DNA, N6-methyladenine is rare — 25 parts per million — but confined to certain parts of the genome. Eliminating DMAD in flies means N6-methyladenine abundance shoots up, and the purified DMAD protein can remove N6-methyladenine from DNA in a test tube.
Jin says that one significant finding in the current paper is that Polycomb proteins, which repress gene transcription in both flies and humans, could be potential “readers” for N6-methyladenine. N6-methyladenine is present more often on DNA where Polycomb binds, and Polycomb prefers to bind DNA containing N6-methyladenine, the researchers showed.
Research article: Active N6-Methyladenine Demethylation by DMAD Regulates Gene Expression by Coordinating with Polycomb Protein in Neurons

